This post mirrors the abstract accepted to the Orthopedic Research Society Annual Meeting 2019, which can be found here. You can check out the poster that I presented here.
Authors
Rehaan M. Bhimani1, Claire J. Watson1, A. Murat Maga2, Ronald Y. Kwon1
1University of Washington, 2Seattle Children’s Research Institute
Introduction
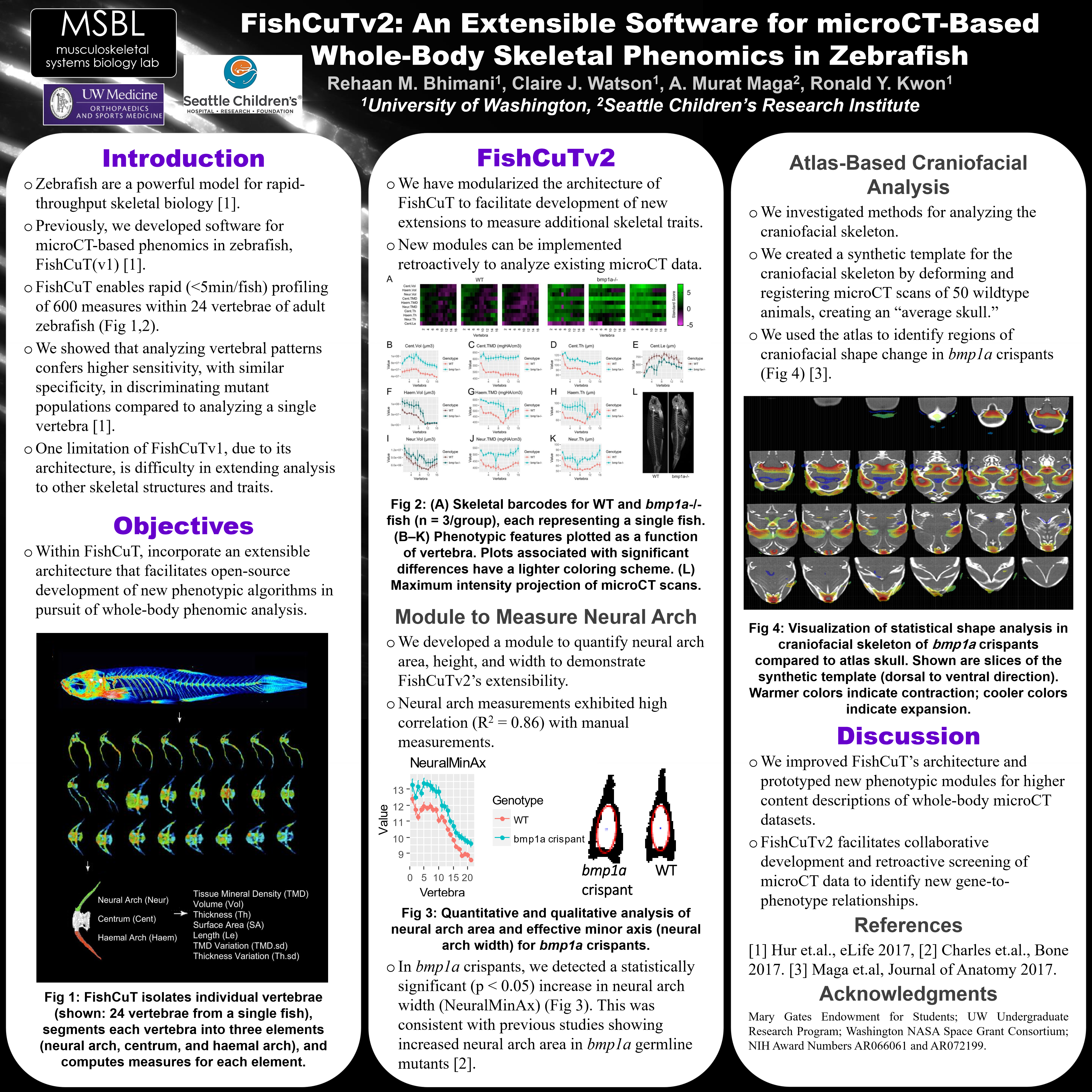
While technological advances have revolutionized our ability to sequence genomes, our ability to characterize vertebrate phenomes remains limited. Zebrafish are a powerful model for rapid-throughput biology, and ideal to understand the genetic architecture of skeletal health through rapid gene-to-phenome mapping. Previously, we developed software for microCT-based phenomics, FishCuT, which enables rapid (<5min/animal) profiling of hundreds of phenotypic measures comprised of morphological and densitometric traits at a large number of sites within the axial skeleton of adult zebrafish (Fig 1A) [1]. We showed that analyzing vertebral patterns conferred higher sensitivity, with similar specificity, in discriminating mutant populations compared to analyzing single vertebrae in isolation [1]. Here, we introduce FishCuTv2, which employs an extensible architecture that facilitates open-source development of new phenotypic algorithms in pursuit of truly whole-body skeletal phenomic analysis. We develop a new algorithm for automatic measurement of neural arch area, a measure of bone modeling in zebrafish [2], and use it to retroactively identify new gene-to-phenotype relationships in CRISPR-edited somatic mutants for the Osteogenesis Imperfecta (OI)-associated gene bmp1a. Finally, we provide early evidence of the utility of atlas-based methods to facilitate microCT-based phenomics in the adult craniofacial skeleton. METHODS: This study was conducted using ~3 month old sham control zebrafish (n=15) and CRISPR edited F0 bmp1a “crispant” zebrafish (n=15), which were previously shown to closely reflect the skeletal phenotypes in bmp1a germline mutants [3]. FishCuT software architecture was reorganized into a hierarchical structure with discrete phenotypic modules, thus allowing for addition of new modules to increase phenotypic functionality. A new module was established to automatically measure phenotypic characteristics of neural arch space (defined as the empty space enclosed by the neural arch and the top of the centrum): effective major axis length, effective minor axis length, and area. For craniofacial analysis, we used ANTs [4] and scan data to create a synthetic template for the adult zebrafish craniofacial skeleton. A statistical shape analysis of the craniofacial skeleton was performed using tensor-based morphometry [5].
Figure 1. (A) Schematic of workflow for phenomic profiling. FishCuTv2 isolates individual vertebrae, segments each vertebra into three elements, and computes traits on each element. (B) Neural arch space effective minor axis length plotted as a function of vertebra. (B’) Image of binarized and inverted neural arch with automatically fitted ellipse used to determine length of effective major and minor axes. Image taken from 6 th vertebra of similarly sized fish: bmp1a crispant on the left and sham control on the right. (C) Visual representation of statistical shape analysis in craniofacial skeleton of bmp1a crispants compared to atlas skull. Images depict slices along the dorsoventral axis. Warmer colors indicate contraction; cooler colors indicate expansion.
Results
Neural arch space measurements exhibited a high correlation (R2=0.86) to measurements made by hand. In bmp1a crispants, we detected a statistically significant (p<0.05) increase in neural arch space area, which is consistent with the findings in [2] for bmp1a germline mutants. While minor axis was significantly different (Figure 1.B), major axis was not, indicating that the increased area of the neural arch space is due to a widening of the neural arch space and not lengthening (Figure 1.B’). For craniofacial analysis, we observed convergence during template building, with only a few iterations producing a template that was sufficiently detailed. The statistical shape analysis revealed contractions and expansions of individual voxels in bmp1a crispants. Morphological changes were characterized by focal regions of contraction and expansion, particularly adjacent to the jaw and orbital regions (Figure 1.C). DISCUSSION: In zebrafish, neural arch area expands throughout life due to coupled activity between osteoclasts on the inner surface, and osteoblasts on the outer surface. The increased neural arch area suggests increased bone modeling arising from loss of function of Bmp1a. The neural arch measurement module serves as a prototype for the development of new phenotypic modules that enable increasingly granular descriptions of whole-body, high-resolution microCT datasets. Because of the new software modularization, developers who wish to create new modules do not need to know the inner workings of FishCuTv2, but rather only the available input variables and necessary output data structures for measurement modules. This facilitates collaborative development, and retroactive screening of microCT data archives for new gene-to-phenotype relationships. Due to its complex morphology and prominence of bone fusions, quantitative analysis of craniofacial structures in the adult zebrafish skeleton is challenging. We found that a detailed synthetic template of the WT craniofacial skeleton could be computed, and thus, may facilitate atlas-based segmentation and other analytical methods. In statistical shape analysis, we found significant craniofacial shape changes in bmp1a crispants that were not obvious by manual inspection. Craniofacial features have been suggested to stratify in OI types, and thus, these studies suggest that microCT-based craniofacial analysis in zebrafish may aid in the study of this stratification. Taken together, these studies establish extensible software and algorithmic methods that take a significant step towards true whole-body skeletal phenomic analysis in zebrafish and rapid-throughput gene-to-phenome mapping.
SIGNIFICANCE/CLINICAL RELEVANCE
Zebrafish are an ideal model to understand the genetic basis of adult skeletal health through rapid gene-to-phenome mapping. Here, we develop software architecture and new algorithmic approaches to facilitate in-depth, whole-body skeletal phenotyping in zebrafish, and show how phenomics can reveal relationships relevant to the heritable skeletal disorder OI.
References
[1] Hur et.al., eLife 2017. [2] Charles et.al., Bone 2017. [3] Watson et.al., ORS 2018. [4] Avants et.al, Front Neuroinform 2014. [5] Maga et.al, Journal of Anatomy 2017.
ACKNOWLEDGEMENTS
This project was supported by NIH grants AR066061 and AR072199, the University of Washington Mary Gates Endowment for Students, the University of Washington Undergraduate Research Program, and the Washington NASA Space Grant Consortium.
Poster Presented at Orthopedic Research Society Annual Meeting 2019